Leukocyte Recognition Using EM-Algorithm
Mario Chirinos Colunga, Oscar Sánchez Siordia, and Stephen J. Maybank
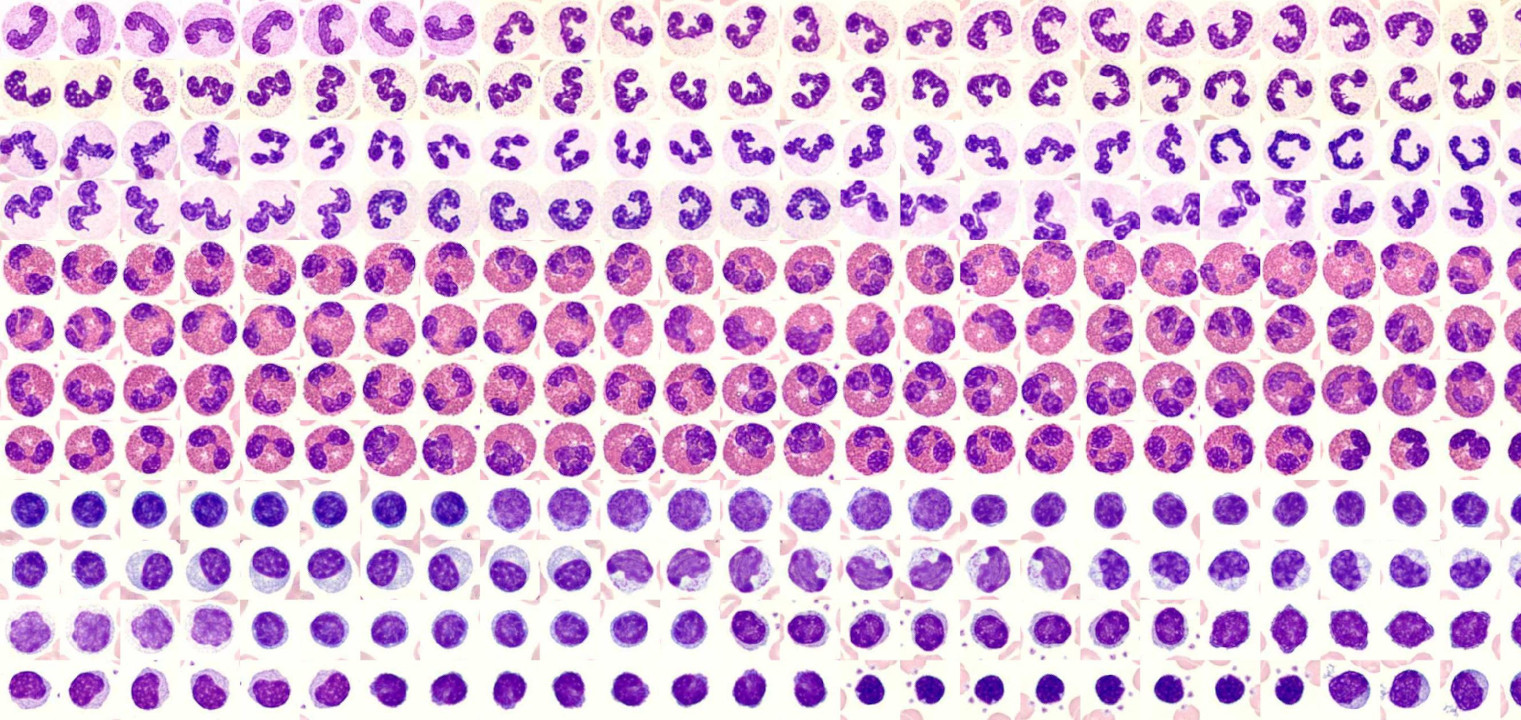
Method for classifying images of blood cells. Three different classes of cells are used: Band Neutrophils,
Eosinophils and Lymphocytes. The image pattern is projected down to a lower dimensional sub space using PCA; the probability density function for each class is modeled with a Gaussian mixture using the EM-Algorithm. A new cell image is classified using the maximum a posteriori decision rule.
Blood Smears
The leukocyte images used for this work were acquired from “Hemosurf -an Interactive Hematology Atlas” for research proposes. The images were taken with a Sony DXC-3000A 3CCD
video camera and a Zeiss Axioskop microscope using a 100X objective and a 10x eyepiece.
Segmentation and Labeling

A “ground truth” for each one of the images was created manually by labeling the regions of interest: nucleus, cytoplasm and background. Leukocytes are extracted from the images by centering a square window at the center of mass of the cytoplasm.
Feature Extraction and Selection

The data set is projected down into a lower dimensional space using PCA and the first 7 principal components, the number of principal component used was found empirically which is the maximum number of principal components that allow to have non-singular covariance matrices for all the mixture components.
Supervised Learning
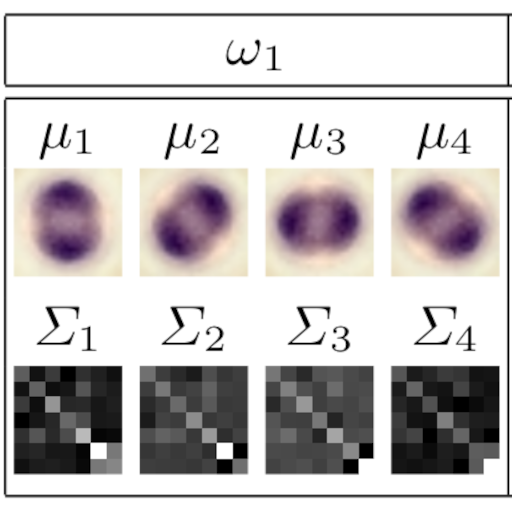
The class conditional density function p(x|ωc) for each leukocyte class is modeled using a mixture of Gaussian densities. To find the parameters of each mixture, the mean vectors and the covariance matrices the EM-Algorithm is applied independently to each class
Classification
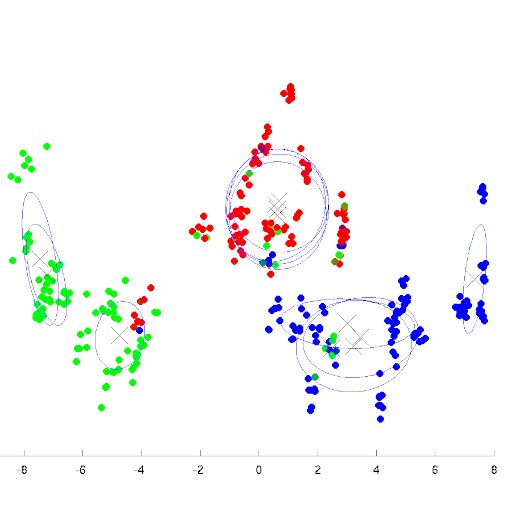
A new leukocyte image is classified using the maximum posteriory rule, the posterior probabilities are found using the Bayes’ theorem.
Research Paper

Leukocyte Recognition Using EM-Algorithm
8th Mexican International Conference on Artificial Intelligence, Guanajuato, México, November 9-13, 2009 Proceedings






